

About me
srkobakian
Master of Philosophy in Statistics (QUT)
Bachelor of Commerce, Bachelor of Economics
I live in Melbourne, Australia
I spent most of my masters researching map displays used in Cancer Atlases. To explore the use of alternative displays, and select one that would work for Australia.
We couldn't find one that would really improve the choropleth map display

Ask me a question
Use the chat
Answer in discussion time
Now there may have been some terms I used that are unfamiliar to you. And that is great because it means I may get to teach you something today. So If you have a question during the talk, use the chat to send it through, we may answer it as the presentation continues but if we don't i'll address it in the discussion time.
What can you do with ggplot2?
What can you do with ggplot2?
Use the Grammar of Graphics to build visulisations

What is the geom for mapping?
What is the geom for mapping?
This is not a simple question
There are many types of maps
ggplot2: Elegant Graphics for Data Analysis:

This is not a simple question There are many ways to map
What do we have to work with?
What do we need to plot?
Libraries
library(ggplot2)library(maps)Data
world <- map_data("world")Australia <- world %>% filter(region == "Australia") Australia## long lat group order region subregion## 1 123.5945 -12.42568 133 7115 Australia Ashmore and Cartier Islands## 2 123.5952 -12.43594 133 7116 Australia Ashmore and Cartier Islands## 3 123.5732 -12.43418 133 7117 Australia Ashmore and Cartier Islands## 4 123.5725 -12.42393 133 7118 Australia Ashmore and Cartier Islands## 5 123.5945 -12.42568 133 7119 Australia Ashmore and Cartier Islands## 6 158.8788 -54.70976 139 7267 Australia Macquarie IslandMapping Data
ggplot(Australia) + geom_point(aes(x = long, y = lat))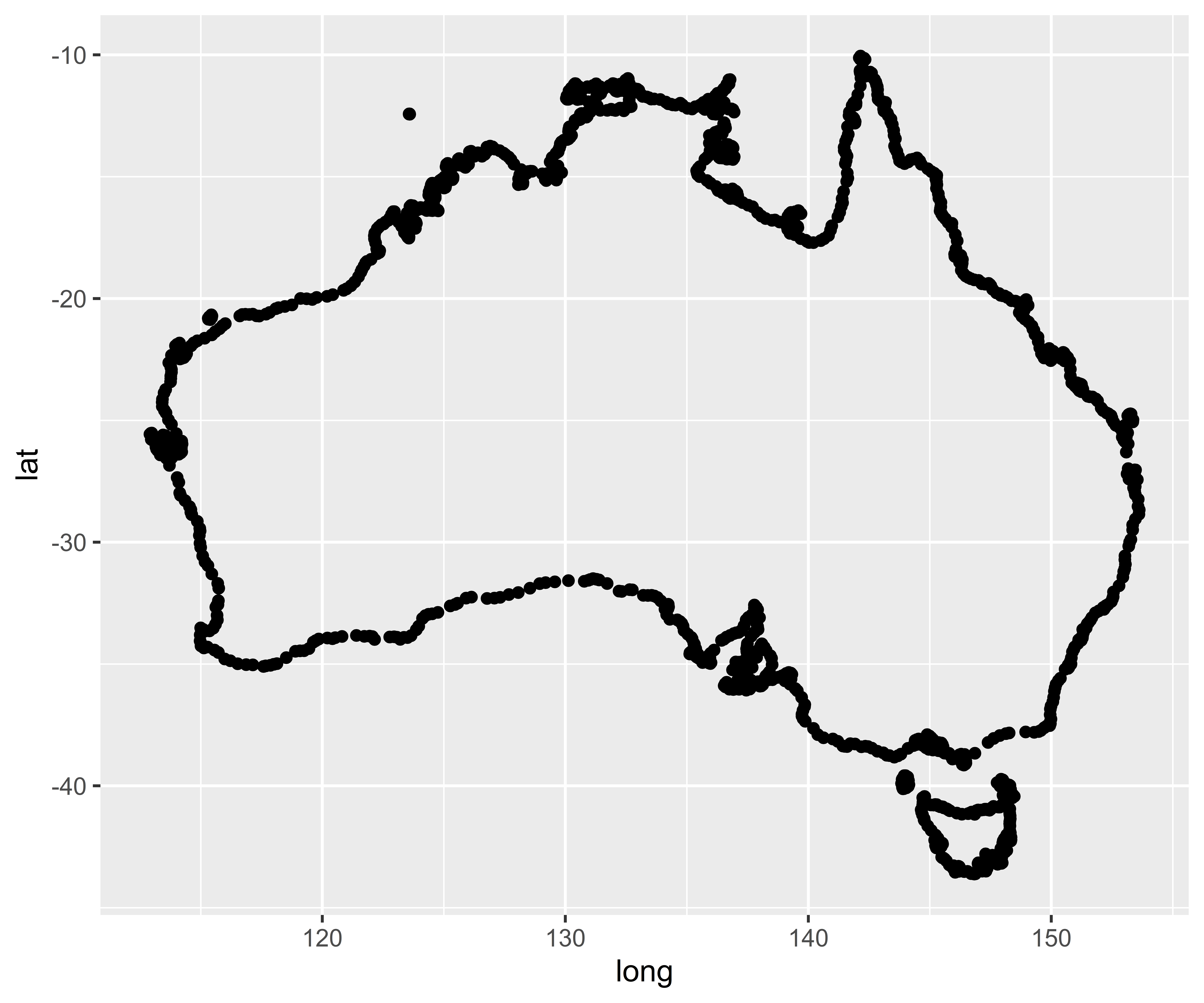
ggplot(Australia) + geom_line(aes(x = long, y = lat))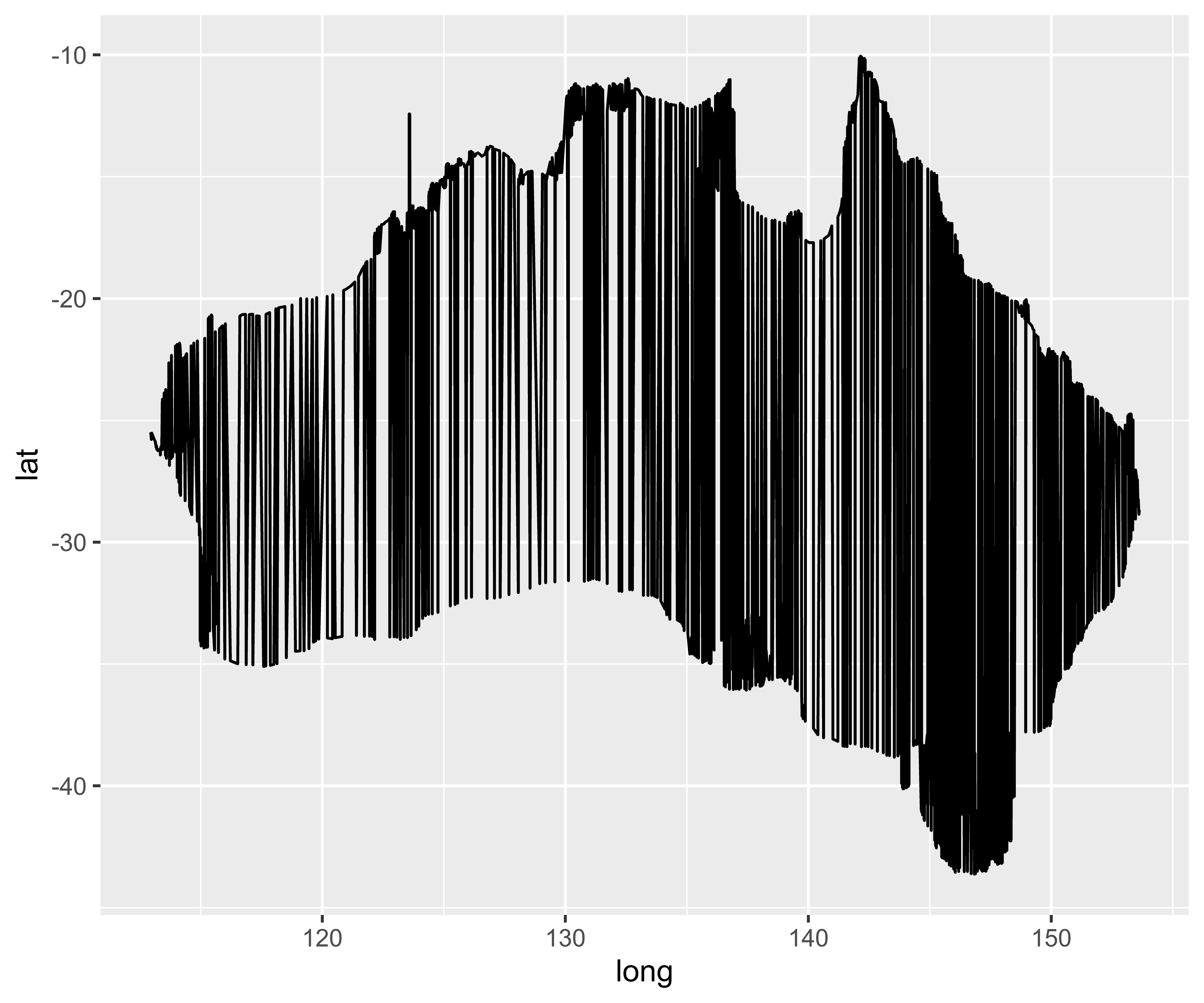
These points are what we need for a map. But it is about how you use them.
Polygon Mapping
ggplot(Australia) + geom_polygon(aes(x = long, y = lat, group = group))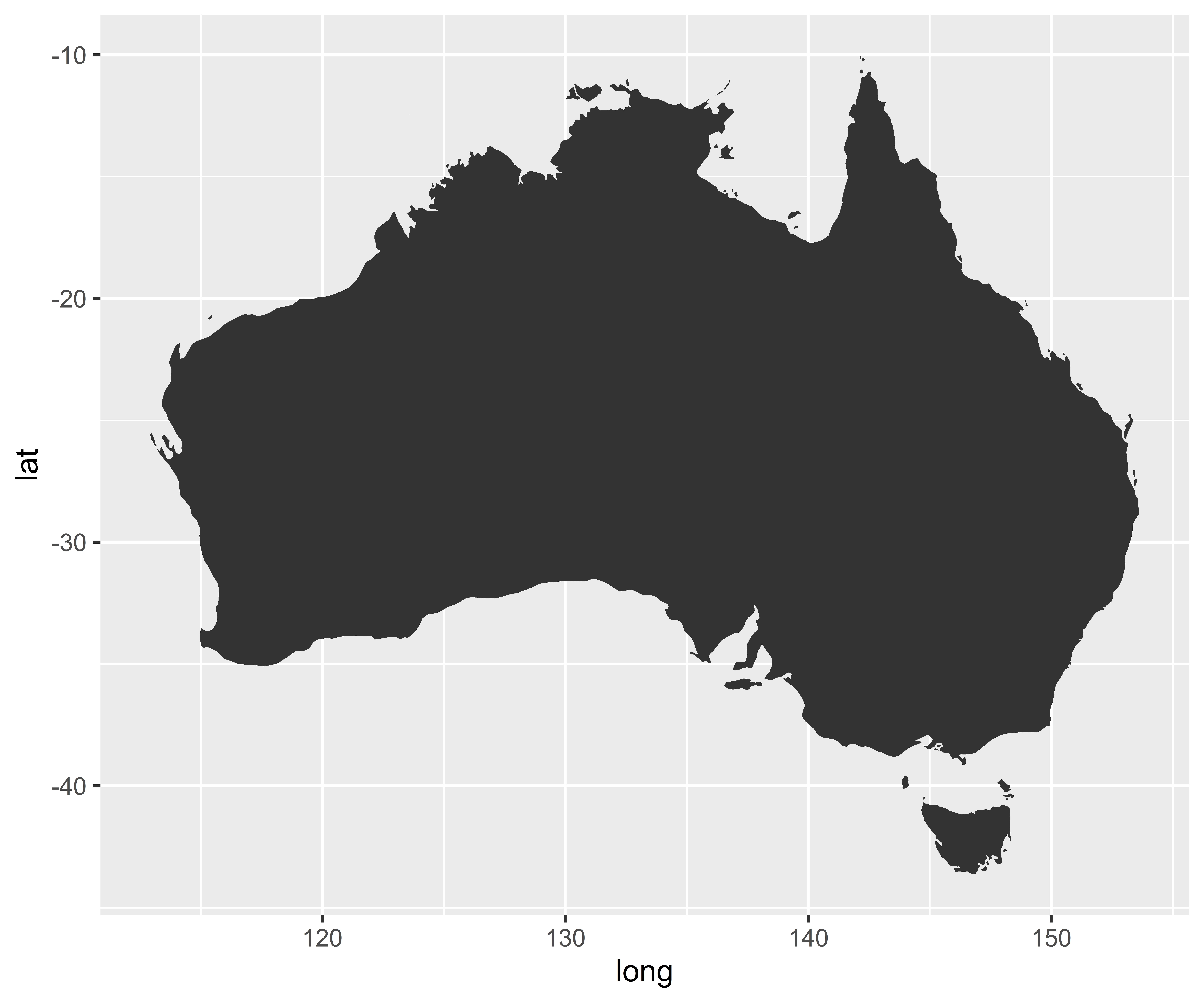
This is a great start
- It looks like Australia!
- Borders are clear
- Islands are also clear
How can we improve this?
Polygons are very similar to paths (as drawn by geom_path()) except that the start and end points are connected and the inside is coloured by fill.
SF system
Simple Features
Key difference:
geometry list-column
Simple Features
Key difference:
geometry list-column
Converts many, many rows
into a single row per area
Geometries fit together,
like puzzle pieces
ozmaps package
library(sf)library(ozmaps)sf_oz <- ozmap_data("country")sf_oz %>% kable()| NAME | geometry |
|---|---|
| Australia | MULTIPOLYGON (((144.8691 -4... |
ggplot(sf_oz) + geom_sf()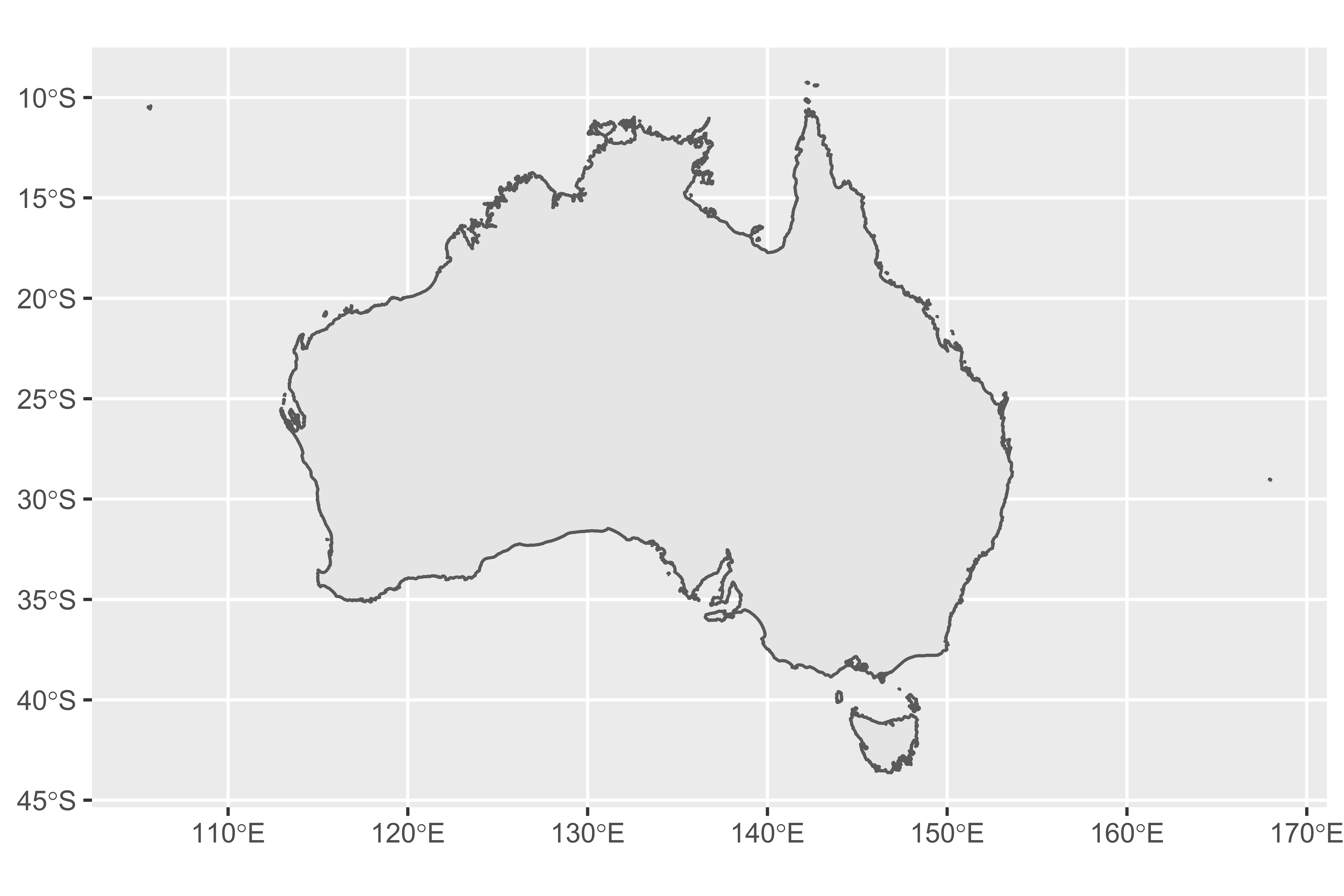
| NAME | geometry |
|---|---|
| New South Wales | MULTIPOLYGON (((150.7016 -3... |
| Victoria | MULTIPOLYGON (((146.6196 -3... |
| Queensland | MULTIPOLYGON (((148.8473 -2... |
| South Australia | MULTIPOLYGON (((137.3481 -3... |
| Western Australia | MULTIPOLYGON (((126.3868 -1... |
| Tasmania | MULTIPOLYGON (((147.8397 -4... |
| Northern Territory | MULTIPOLYGON (((136.3669 -1... |
| Australian Capital Territory | MULTIPOLYGON (((149.2317 -3... |
| Other Territories | MULTIPOLYGON (((167.9333 -2... |
sf_states <- ozmap_data("states")ggplot(sf_states) + geom_sf()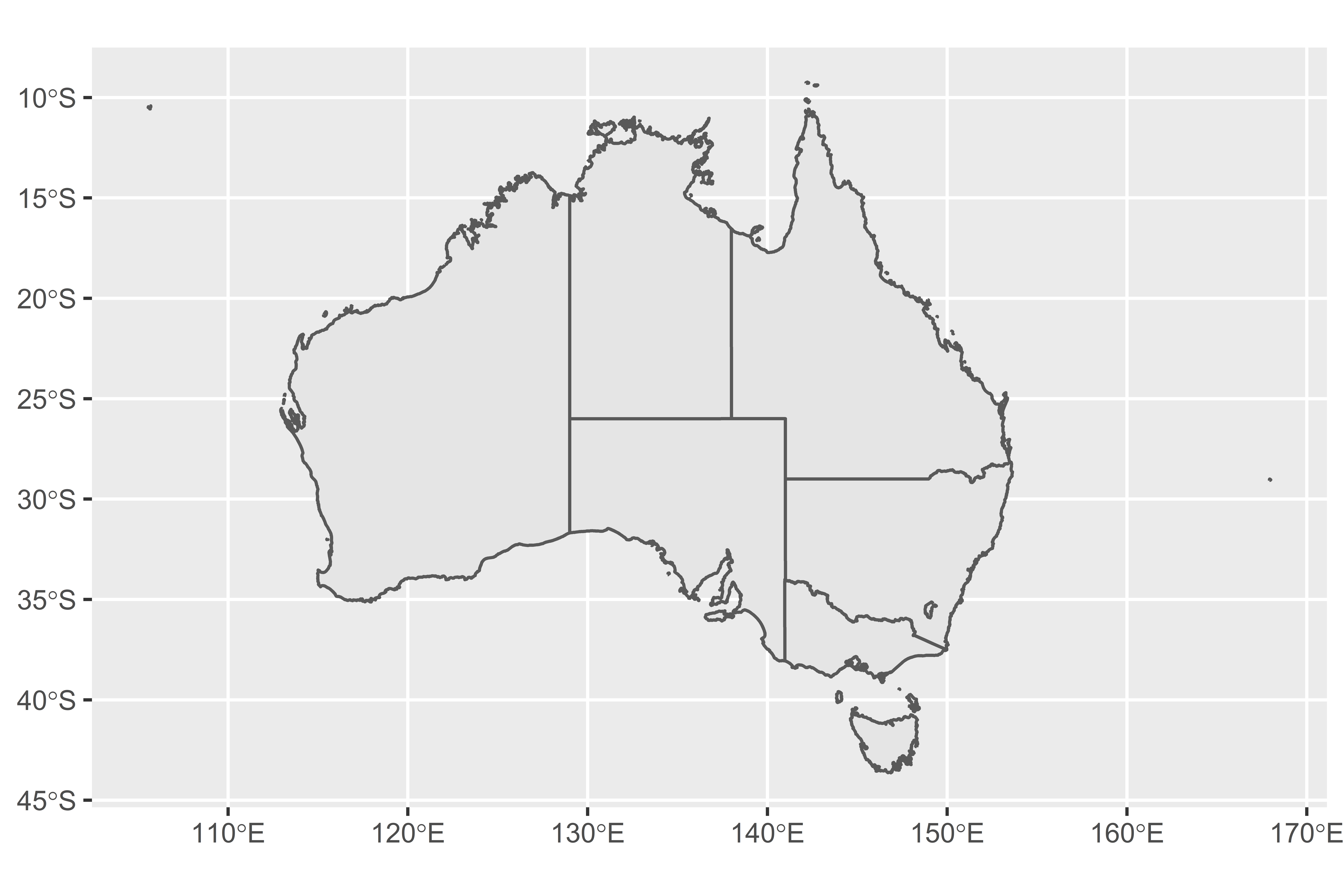
Creating Choropleth maps
A choropleth maps fills regions on the maps according to their data value
Creating Choropleth maps
A choropleth maps fills regions on the maps according to their data value
We need two things:
Map and Data
Data: COVID LIVE Australia
library(rvest)library(polite)covid_url <- "https://covidlive.com.au/report/cases"covid_data <- bow(covid_url) %>% scrape() %>% html_table() %>% purrr::pluck(2) %>% as_tibble()covid_data## # A tibble: 9 x 5## STATE CASES OSEAS VAR NET## <chr> <chr> <int> <lgl> <int>## 1 Victoria 20,479 NA NA 0## 2 NSW 5,150 1 NA 1## 3 Queensland 1,323 2 NA 2## 4 WA 912 1 NA 1## 5 SA 610 NA NA 0## 6 Tasmania 234 NA NA 0## 7 ACT 118 NA NA 0## 8 NT 104 NA NA 0## 9 Australia 28,930 4 NA 4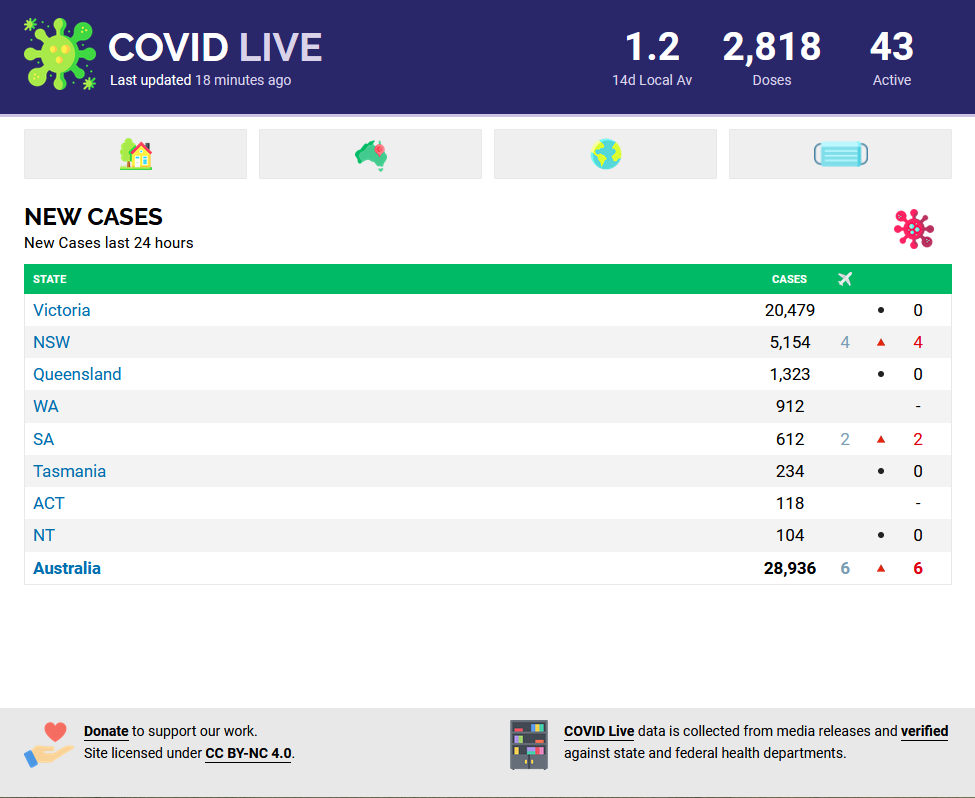
Data
| STATE | CASES | OSEAS | NET |
|---|---|---|---|
| Victoria | 20,479 | NA | 0 |
| NSW | 5,150 | 1 | 1 |
| Queensland | 1,323 | 2 | 2 |
| WA | 912 | 1 | 1 |
| SA | 610 | NA | 0 |
| Tasmania | 234 | NA | 0 |
| ACT | 118 | NA | 0 |
| NT | 104 | NA | 0 |
| Australia | 28,930 | 4 | 4 |
Map
| NAME | geometry |
|---|---|
| New South Wales | MULTIPOLYGON (((150.7016 -3... |
| Victoria | MULTIPOLYGON (((146.6196 -3... |
| Queensland | MULTIPOLYGON (((148.8473 -2... |
| South Australia | MULTIPOLYGON (((137.3481 -3... |
| Western Australia | MULTIPOLYGON (((126.3868 -1... |
| Tasmania | MULTIPOLYGON (((147.8397 -4... |
| Northern Territory | MULTIPOLYGON (((136.3669 -1... |
| Australian Capital Territory | MULTIPOLYGON (((149.2317 -3... |
| Other Territories | MULTIPOLYGON (((167.9333 -2... |
To join the Covid data to the map
To join the Covid data to the map
- Expand the abbreviations so the names match
- Convert cases to numeric values
covid_data <- covid_data %>% mutate(STATE = case_when( # replace the abbreviations STATE == "NSW" ~ "New South Wales", STATE == "WA" ~ "Western Australia", STATE == "SA" ~ "South Australia", STATE == "NT" ~ "Northern Territory", STATE == "ACT" ~ "Australian Capital Territory", # keep the rest of the state names TRUE ~ STATE )) %>% mutate(CASES = parse_number(CASES))- Peform a join
covid_states <- left_join(ozmap_states, covid_data, by = c("NAME" = "STATE"))- Remove the "Other Territories"
covid_states <- covid_states %>% filter(!(NAME == "Other Territories"))Data
| STATE | CASES | OSEAS | NET |
|---|---|---|---|
| Victoria | 20479 | NA | 0 |
| New South Wales | 5150 | 1 | 1 |
| Queensland | 1323 | 2 | 2 |
| Western Australia | 912 | 1 | 1 |
| South Australia | 610 | NA | 0 |
| Tasmania | 234 | NA | 0 |
| Australian Capital Territory | 118 | NA | 0 |
| Northern Territory | 104 | NA | 0 |
| Australia | 28930 | 4 | 4 |
Map
| NAME | geometry |
|---|---|
| New South Wales | MULTIPOLYGON (((150.7016 -3... |
| Victoria | MULTIPOLYGON (((146.6196 -3... |
| Queensland | MULTIPOLYGON (((148.8473 -2... |
| South Australia | MULTIPOLYGON (((137.3481 -3... |
| Western Australia | MULTIPOLYGON (((126.3868 -1... |
| Tasmania | MULTIPOLYGON (((147.8397 -4... |
| Northern Territory | MULTIPOLYGON (((136.3669 -1... |
| Australian Capital Territory | MULTIPOLYGON (((149.2317 -3... |
| Other Territories | MULTIPOLYGON (((167.9333 -2... |
Complete map data
| NAME | geometry | CASES | OSEAS | NET |
|---|---|---|---|---|
| New South Wales | MULTIPOLYGON (((150.7016 -3... | 5150 | 1 | 1 |
| Victoria | MULTIPOLYGON (((146.6196 -3... | 20479 | NA | 0 |
| Queensland | MULTIPOLYGON (((148.8473 -2... | 1323 | 2 | 2 |
| South Australia | MULTIPOLYGON (((137.3481 -3... | 610 | NA | 0 |
| Western Australia | MULTIPOLYGON (((126.3868 -1... | 912 | 1 | 1 |
| Tasmania | MULTIPOLYGON (((147.8397 -4... | 234 | NA | 0 |
| Northern Territory | MULTIPOLYGON (((136.3669 -1... | 104 | NA | 0 |
| Australian Capital Territory | MULTIPOLYGON (((149.2317 -3... | 118 | NA | 0 |
Choropleth Map
ggplot(covid_states) + geom_sf(aes(fill = CASES))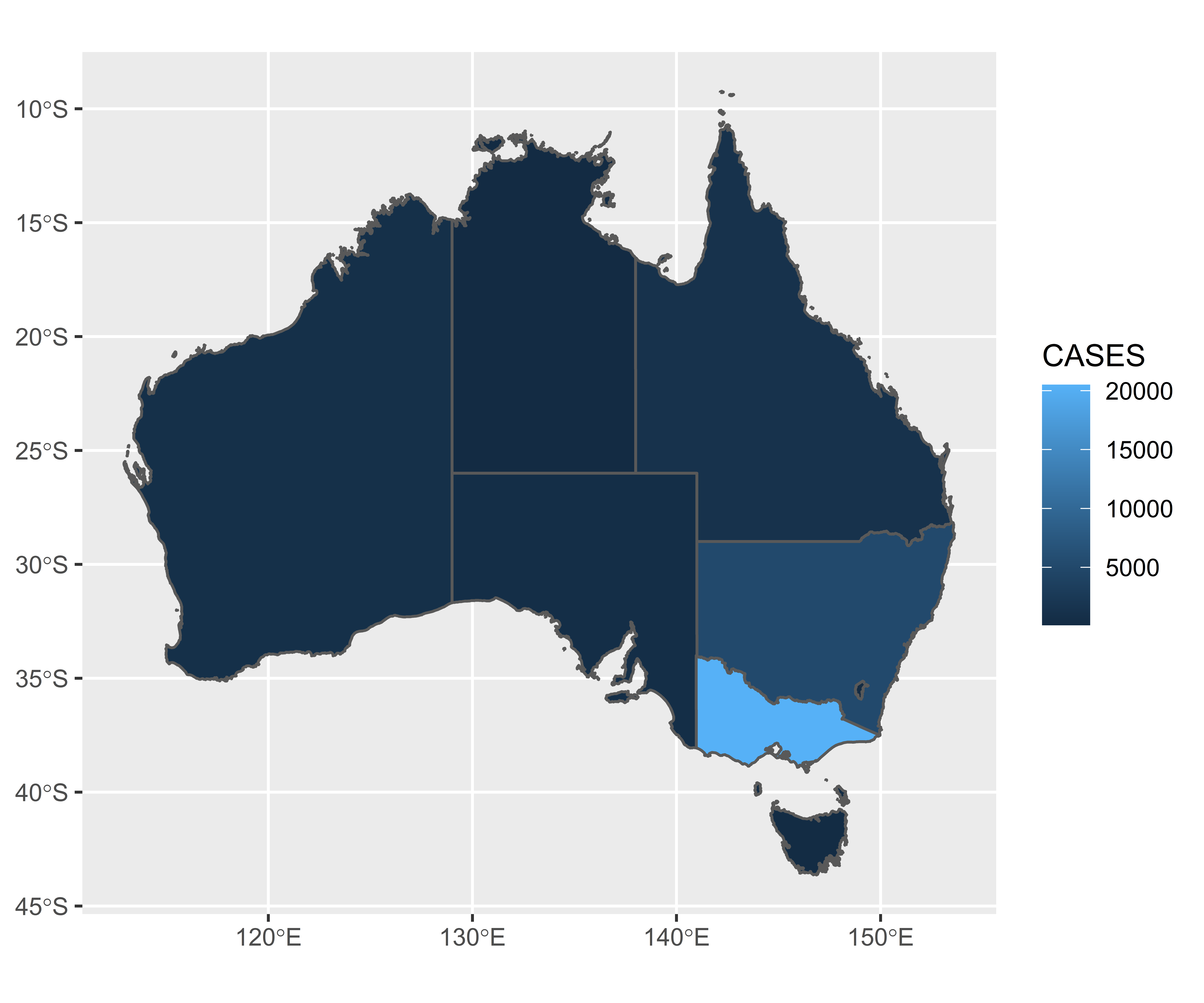
library(ggforce)ggplot(covid_states) + geom_sf(aes(fill = CASES)) + facet_zoom(xy = NAME == "Australian Capital Territory", zoom.size = 0.6)
Off we go!

Covid in Sri Lanka
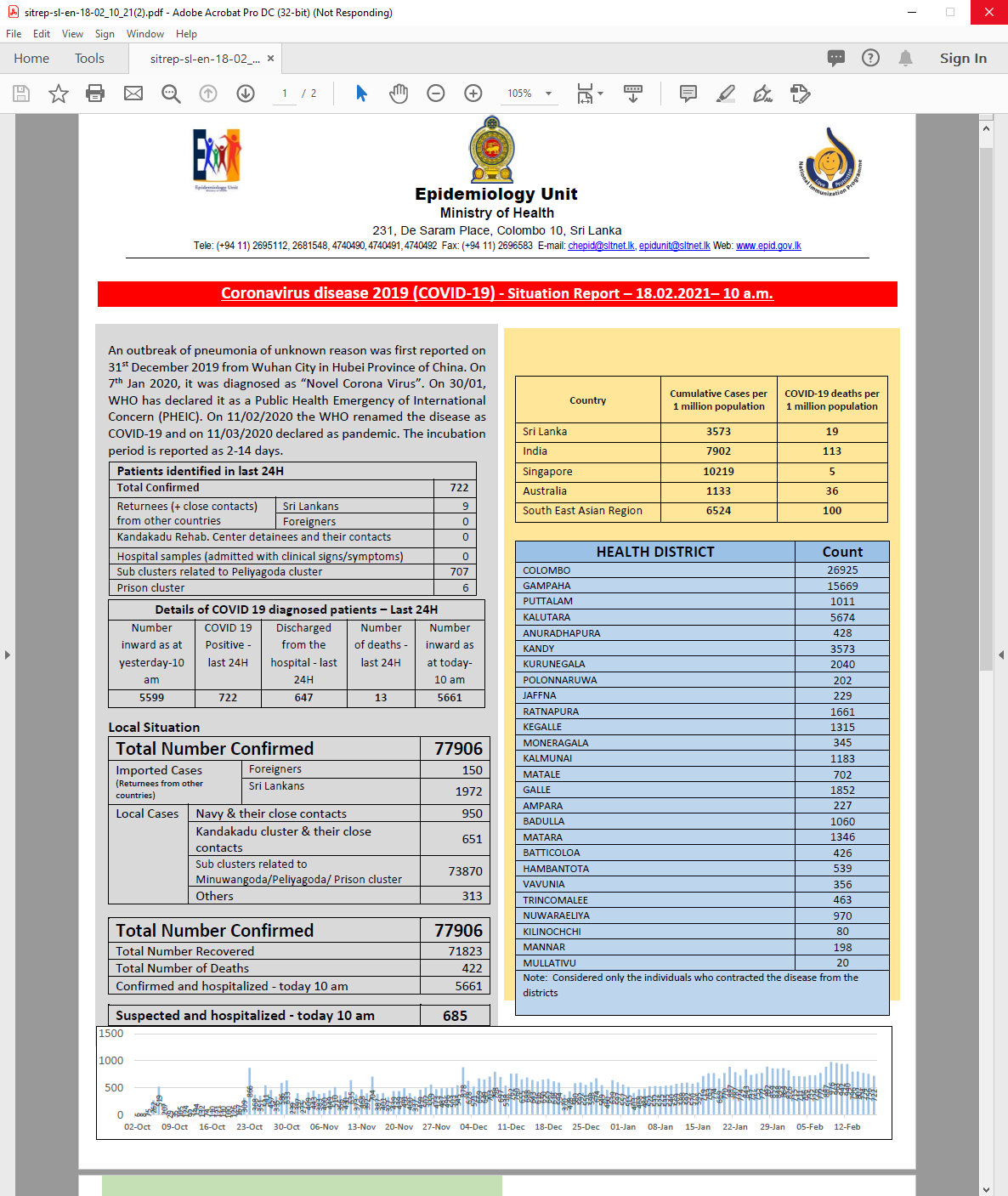
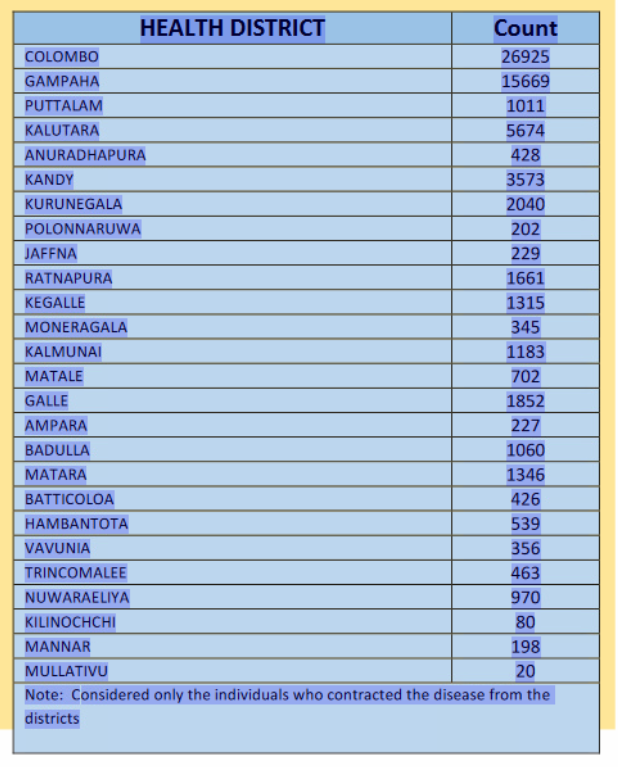
Data: Epidemiology Unit, Ministry of Health
| DISTRICT | Count |
|---|---|
| COLOMBO | 26925 |
| GAMPAHA | 15669 |
| PUTTALAM | 1011 |
| KALUTARA | 5674 |
| ANURADHAPURA | 428 |
| KANDY | 3573 |
| KURUNEGALA | 2040 |
| POLONNARUWA | 202 |
| JAFFNA | 229 |
| RATNAPURA | 1661 |
library(stringr)# https://covid19.gov.lk/wp-content/uploads/2021/02/sitrep-sl-en-18-02_10_21.pdfcovid_sl_table <- c("COLOMBO26925GAMPAHA15669PUTTALAM1011KALUTARA5674ANURADHAPURA428KANDY3573KURUNEGALA2040POLONNARUWA202JAFFNA229RATNAPURA1661KEGALLE1315MONERAGALA345KALMUNAI1183MATALE702GALLE1852AMPARA227BADULLA1060MATARA1346BATTICOLOA426HAMBANTOTA539VAVUNIA356TRINCOMALEE463NUWARAELIYA970KILINOCHCHI80MANNAR198MULLATIVU20")# Extract names of regions, using a regular expression to search for lettersdistricts <- covid_sl_table %>% str_extract_all(string = ., pattern = "[A-Z]*") %>% unlist( ) %>% str_subset(., pattern = "[A-Z]")# Extract number of cases, using a regular expression to search for numberscounts <- covid_sl_table %>% str_extract_all(string = ., pattern = "[0-9]*") %>% unlist() %>% str_subset(., pattern = "[0-9]")# Combine two sets of data to create a tablecovid_sl <- tibble(DISTRICT = districts, Count = counts) %>% mutate(Count = parse_number(Count))Map data for Sri Lanka
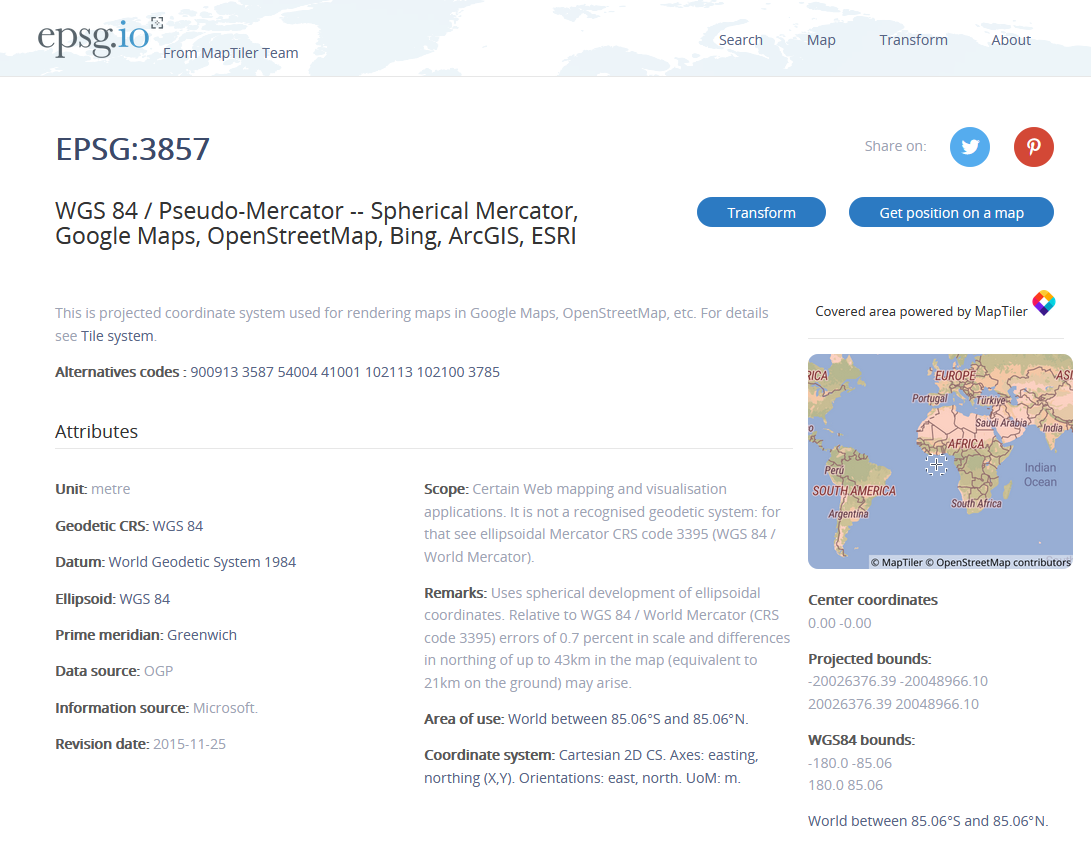
# Select the level 2 districts (adm2)sf_sl <- read_sf("data/lka_adm_slsd_20200305_shp/lka_admbnda_adm2_slsd_20200305.shp") %>% select(ADM2_EN, geometry)sf_sl## Simple feature collection with 26 features and 1 field## geometry type: MULTIPOLYGON## dimension: XY## bbox: xmin: 79.51916 ymin: 5.918297 xmax: 81.8772 ymax: 9.835718## geographic CRS: WGS 84## # A tibble: 26 x 2## ADM2_EN geometry## <chr> <MULTIPOLYGON [°]>## 1 [unknown] (((79.80303 7.8503, 79.80333 7.849463, 79.80333 7.849463, 79.803~## 2 Ampara (((81.23892 7.738495, 81.23903 7.73846, 81.23912 7.738474, 81.23~## 3 Anuradhapu~ (((80.79126 8.918771, 80.79164 8.918763, 80.79209 8.91885, 80.79~## 4 Badulla (((80.9806 7.61768, 80.98104 7.617651, 80.98152 7.617694, 80.982~## 5 Batticaloa (((81.76936 7.536074, 81.76918 7.536048, 81.76907 7.536107, 81.7~## 6 Colombo (((79.87696 6.98085, 79.87805 6.98068, 79.87808 6.980692, 79.878~## 7 Galle (((80.09646 6.132867, 80.09638 6.132866, 80.0963 6.132871, 80.09~## 8 Gampaha (((80.14677 7.312115, 80.14689 7.311824, 80.1469 7.311761, 80.14~## 9 Hambantota (((81.61401 6.572167, 81.61425 6.572167, 81.61462 6.572185, 81.6~## 10 Jaffna (((79.52615 9.383134, 79.52537 9.383026, 79.5245 9.383076, 79.52~## # ... with 16 more rows# Choose a projection from epsg.io:sf_sl <- sf_sl %>% st_transform(5235)Data
| DISTRICT | Count |
|---|---|
| COLOMBO | 26925 |
| GAMPAHA | 15669 |
| PUTTALAM | 1011 |
| KALUTARA | 5674 |
| ANURADHAPURA | 428 |
| KANDY | 3573 |
| KURUNEGALA | 2040 |
| POLONNARUWA | 202 |
| JAFFNA | 229 |
| RATNAPURA | 1661 |
Map
| ADM2_EN | geometry |
|---|---|
| [unknown] | MULTIPOLYGON (((392952.6 59... |
| Ampara | MULTIPOLYGON (((551311.8 58... |
| Anuradhapura | MULTIPOLYGON (((501928.5 71... |
| Badulla | MULTIPOLYGON (((522825 5682... |
| Batticaloa | MULTIPOLYGON (((609877 5593... |
| Colombo | MULTIPOLYGON (((400911.5 49... |
| Galle | MULTIPOLYGON (((425038.8 40... |
| Gampaha | MULTIPOLYGON (((430775.7 53... |
| Hambantota | MULTIPOLYGON (((592923.3 45... |
| Jaffna | MULTIPOLYGON (((362968.6 76... |
A different problem to the Australia set
What is this unknown district?
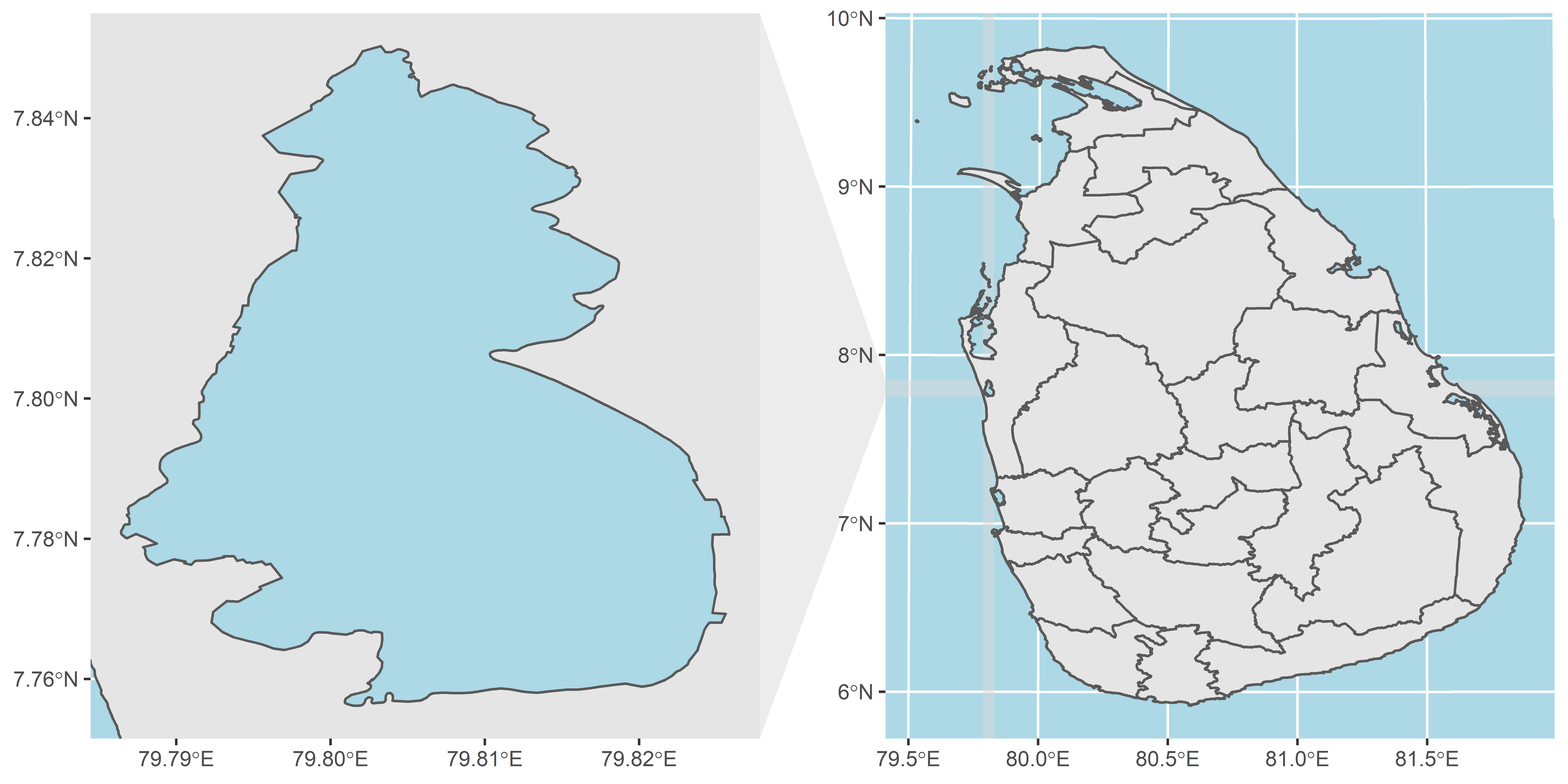
To join the Covid data to the map
To join the Covid data to the map
- Convert the district names to match
covid_sl <- covid_sl %>% # use District names from Covid data mutate(DISTRICT = case_when( DISTRICT == "BATTICOLOA" ~ "BATTICALOA", DISTRICT == "NUWARAELIYA" ~ "NUWARA ELIYA", DISTRICT == "MONERAGALA" ~ "MONARAGALA", DISTRICT == "MULLATIVU" ~ "MULLAITIVU", DISTRICT == "VAVUNIA" ~ "VAVUNIYA", TRUE ~ DISTRICT))sf_sl <- sf_sl %>% mutate(DISTRICT = str_to_upper(ADM2_EN)) %>% select(-ADM2_EN)- Peform a join
covid_districts_sl <- left_join(sf_sl, covid_sl, by = c("DISTRICT"))- Remove the "[unknown]"
covid_districts_sl <- covid_districts_sl %>% filter(!(DISTRICT == "[UNKNOWN]")) %>% filter(!(DISTRICT == "KALMUNAI")) %>% # adjust case numbers # city of Kalmunai in Ampara 1183+227 = 1410 mutate(Count = ifelse(DISTRICT == "AMPARA", 1410, Count))Complete map data
| geometry | DISTRICT | Count |
|---|---|---|
| MULTIPOLYGON (((551311.8 58... | AMPARA | 1410 |
| MULTIPOLYGON (((501928.5 71... | ANURADHAPURA | 428 |
| MULTIPOLYGON (((522825 5682... | BADULLA | 1060 |
| MULTIPOLYGON (((609877 5593... | BATTICALOA | 426 |
| MULTIPOLYGON (((400911.5 49... | COLOMBO | 26925 |
| MULTIPOLYGON (((425038.8 40... | GALLE | 1852 |
| MULTIPOLYGON (((430775.7 53... | GAMPAHA | 15669 |
| MULTIPOLYGON (((592923.3 45... | HAMBANTOTA | 539 |
| MULTIPOLYGON (((362968.6 76... | JAFFNA | 229 |
| MULTIPOLYGON (((411888.2 43... | KALUTARA | 5674 |
| MULTIPOLYGON (((520143.4 55... | KANDY | 3573 |
| MULTIPOLYGON (((432117.3 52... | KEGALLE | 1315 |
| MULTIPOLYGON (((415223 7518... | KILINOCHCHI | 80 |
| MULTIPOLYGON (((425843.2 63... | KURUNEGALA | 2040 |
| MULTIPOLYGON (((405858.7 70... | MANNAR | 198 |
Choropleth Map
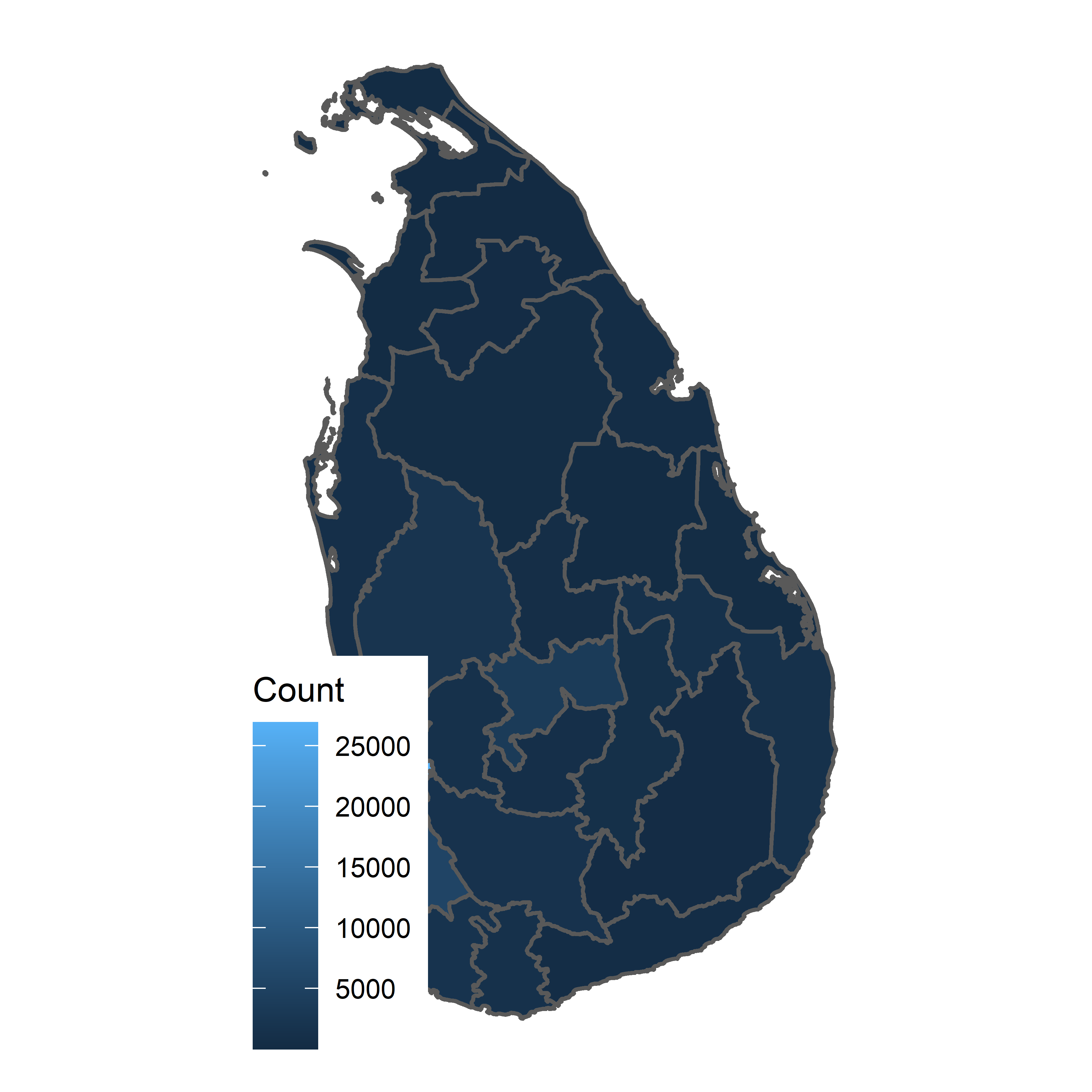
What should we continue to ask ourselves?
What should we continue to ask ourselves?
How can we improve?
Off we go!

We can use projections
We can use projections
A projection converts the 3D globe, into to a 2D representation
Common Projections
Common Projections
EPSG: 4326, World Geodetic System 1984, used in GPS
Common Projections

EPSG: 4326, World Geodetic System 1984, used in GPS
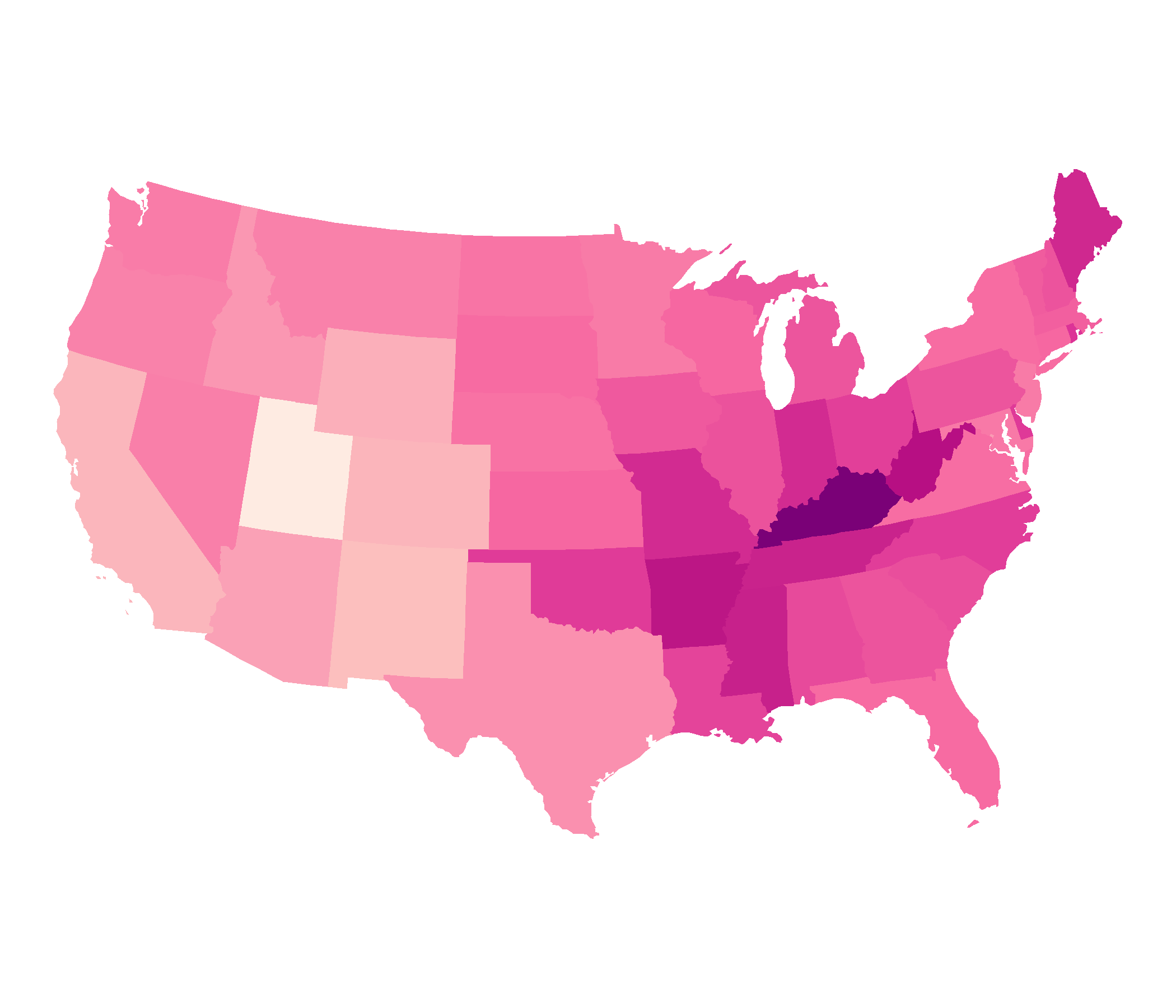
EPSG: 2163, US National Atlas Equal Area, spherical projection
Common Projections

EPSG: 4326, World Geodetic System 1984, used in GPS

EPSG: 2163, US National Atlas Equal Area, spherical projection
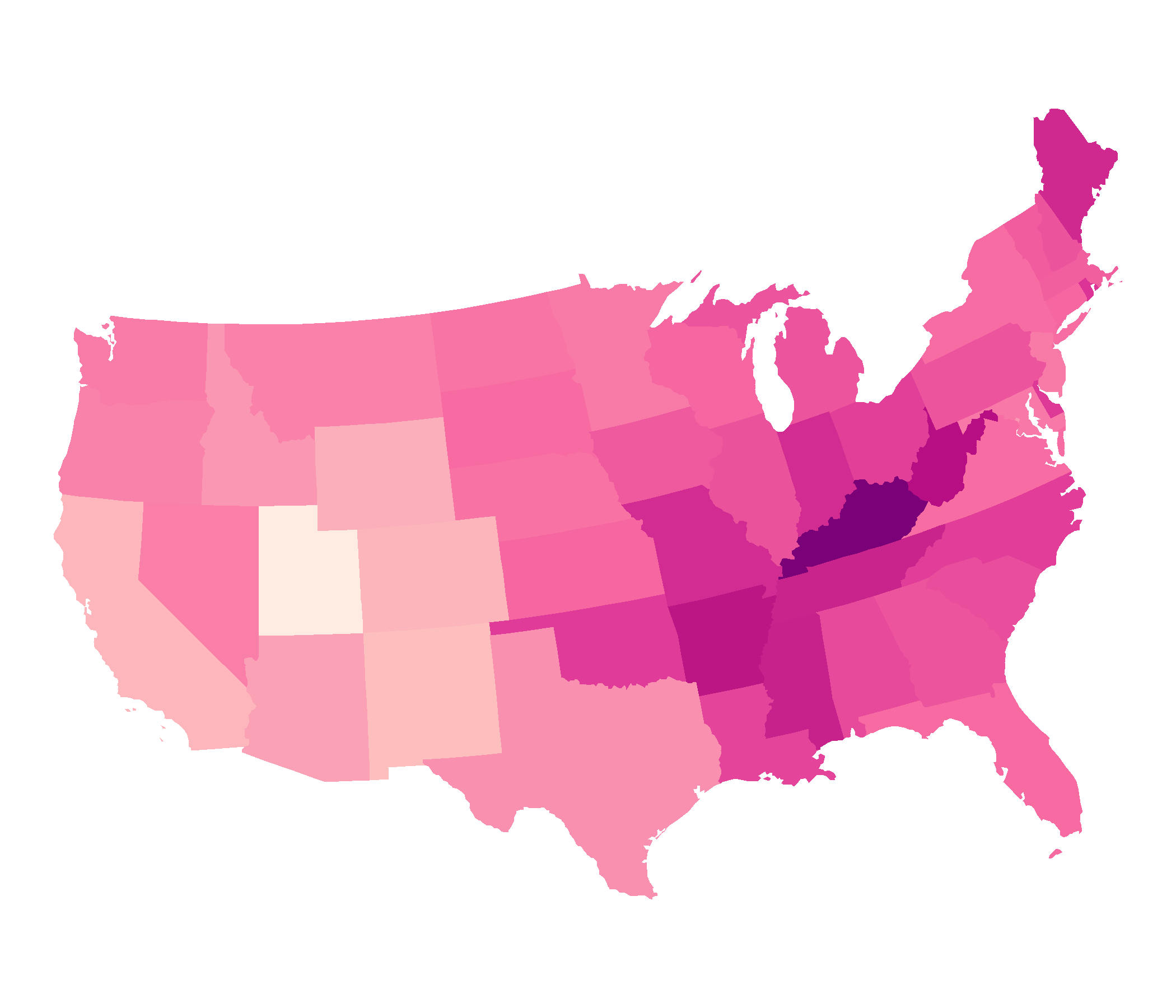
EPSG: 8826, North American Datum 1983
Choropleths
Familiar shapes of areas
Familiar boundary relationships
Identify spatial structures and patterns
However, if we have:
sparsely populated rural areas are easy to see
densely populated inner city areas are geographically small
What are we trying to communicate?
What are the alternatives?
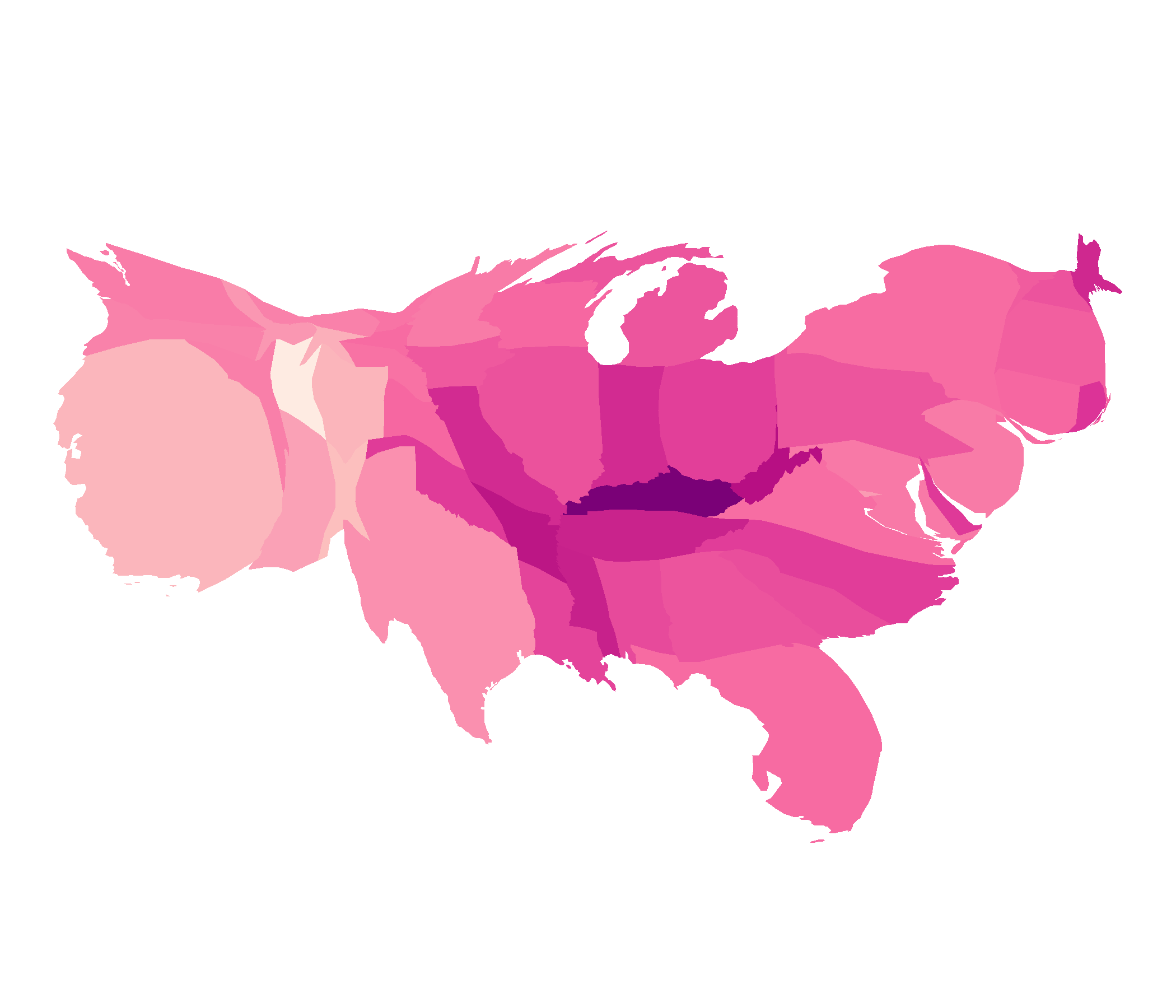
Our paper covers in detail and has lots of examples of world wide cancer atlases
Cartograms
- augment the size, shape, or distance of geographical areas
- sizes areas by the value of a statistic, not earth size area
- account for the population density, reveal hidden spatial patterns
- reducing the visual impact of large areas with small populations
map creators can use white lies to create useful displays by distorting the geometry and suppressing features. [26]. The distortion in an area cartogram accounts for the population density, preventing it from obscuring the spatial patterns [27].
Off we go!

Population
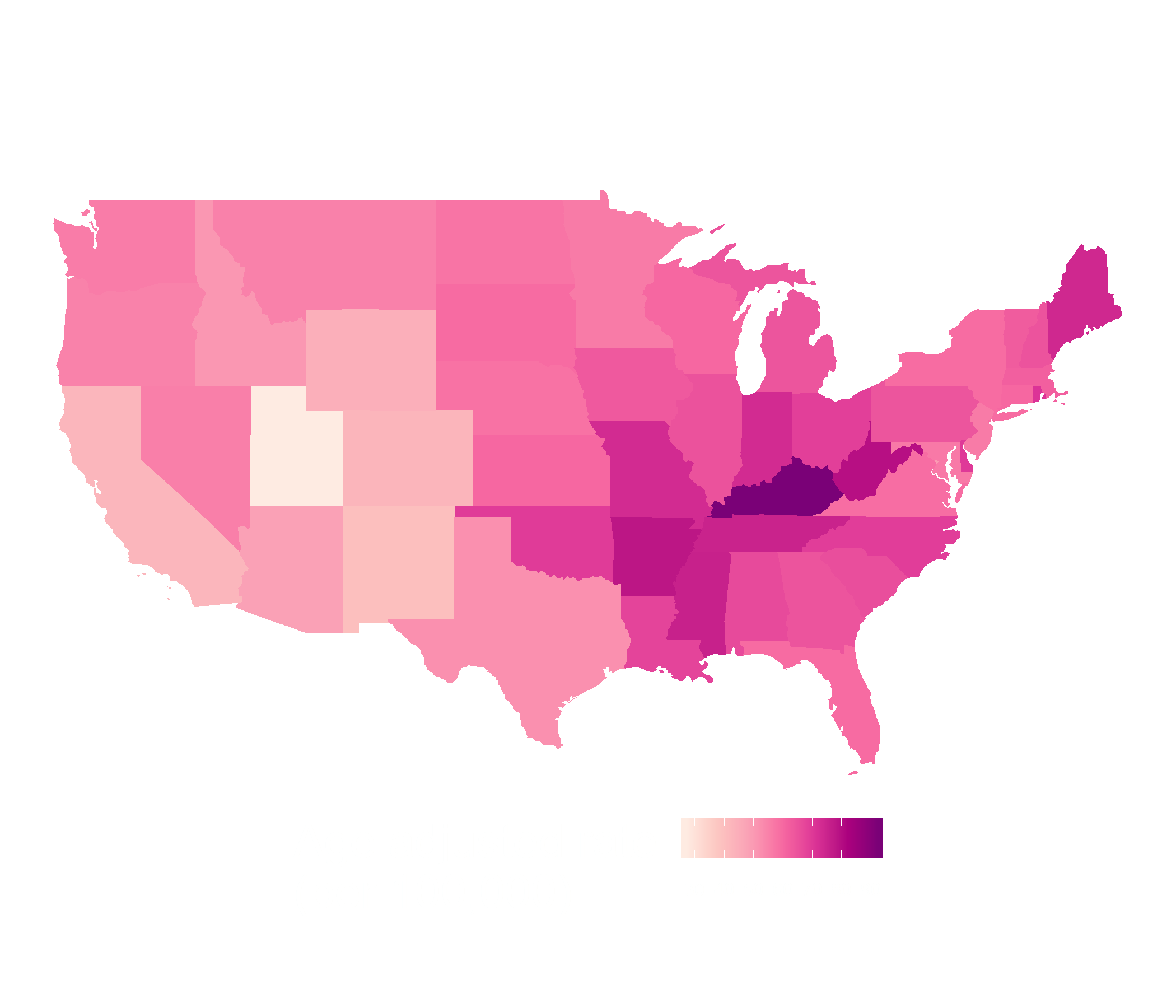
pop_url <- "https://www.citypopulation.de/en/srilanka/prov/admin/"pop_table <- bow(pop_url) %>% scrape() %>% html_table() %>% purrr::pluck(1)pop_data <- pop_table %>% select(DISTRICT = Name, Status, population = `PopulationEstimate2020-07-01`) %>% filter(Status == "District") %>% mutate(DISTRICT = str_to_upper(DISTRICT)) %>% mutate(DISTRICT = ifelse( DISTRICT == "MONERAGALA", "MONARAGALA", DISTRICT)) %>% mutate(population = parse_number(population))covid_districts_sl <- left_join(covid_districts_sl, pop_data)Contiguous Cartogram
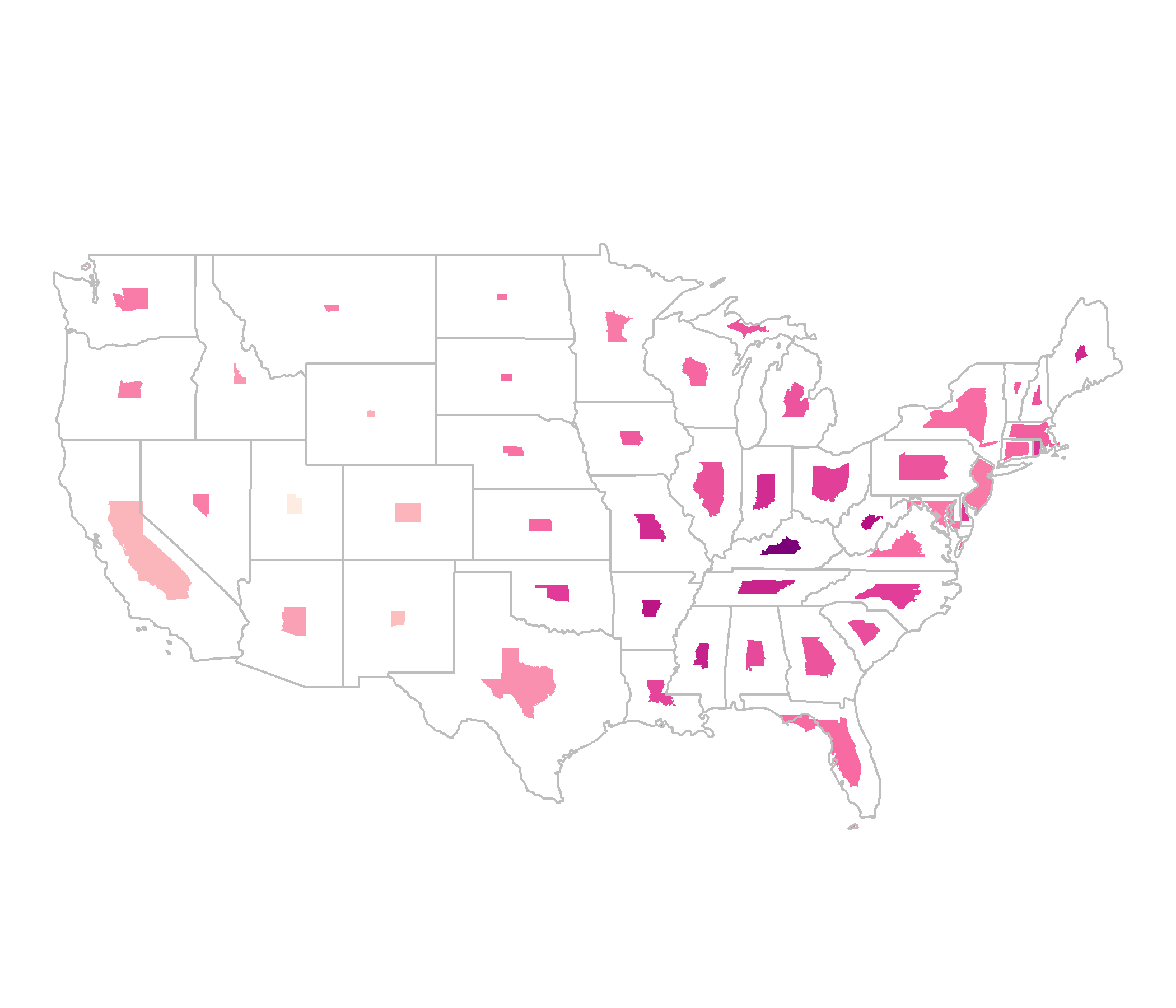
library(cartogram)cont <- cartogram_cont(covid_districts_sl, weight = "population") %>% st_as_sf()ggplot(cont) + geom_sf(aes(fill = Count), colour = NA) + scale_fill_distiller(type = "seq", palette = "RdPu", direction = 1, ) + theme_map() + guides(legend.position = "right")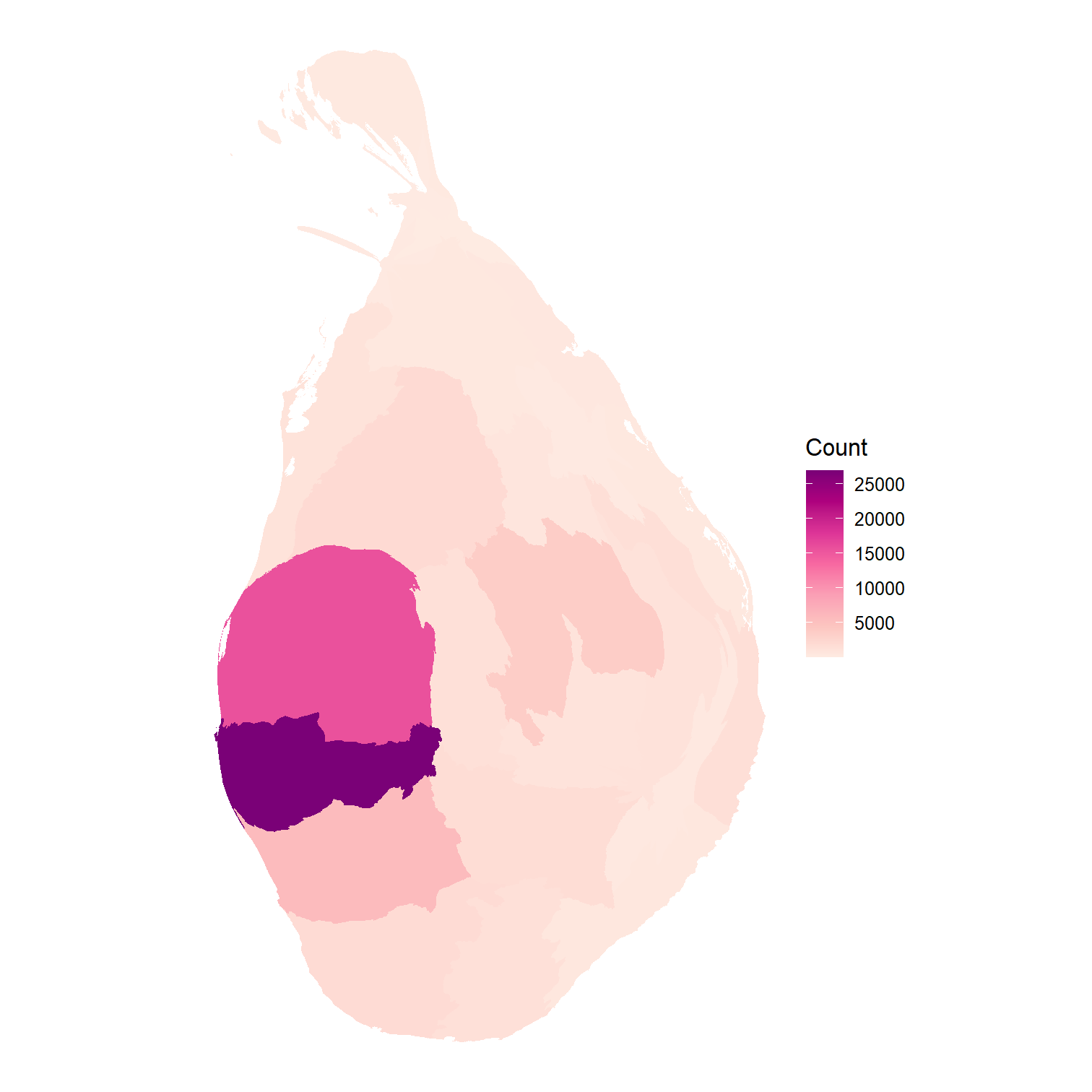
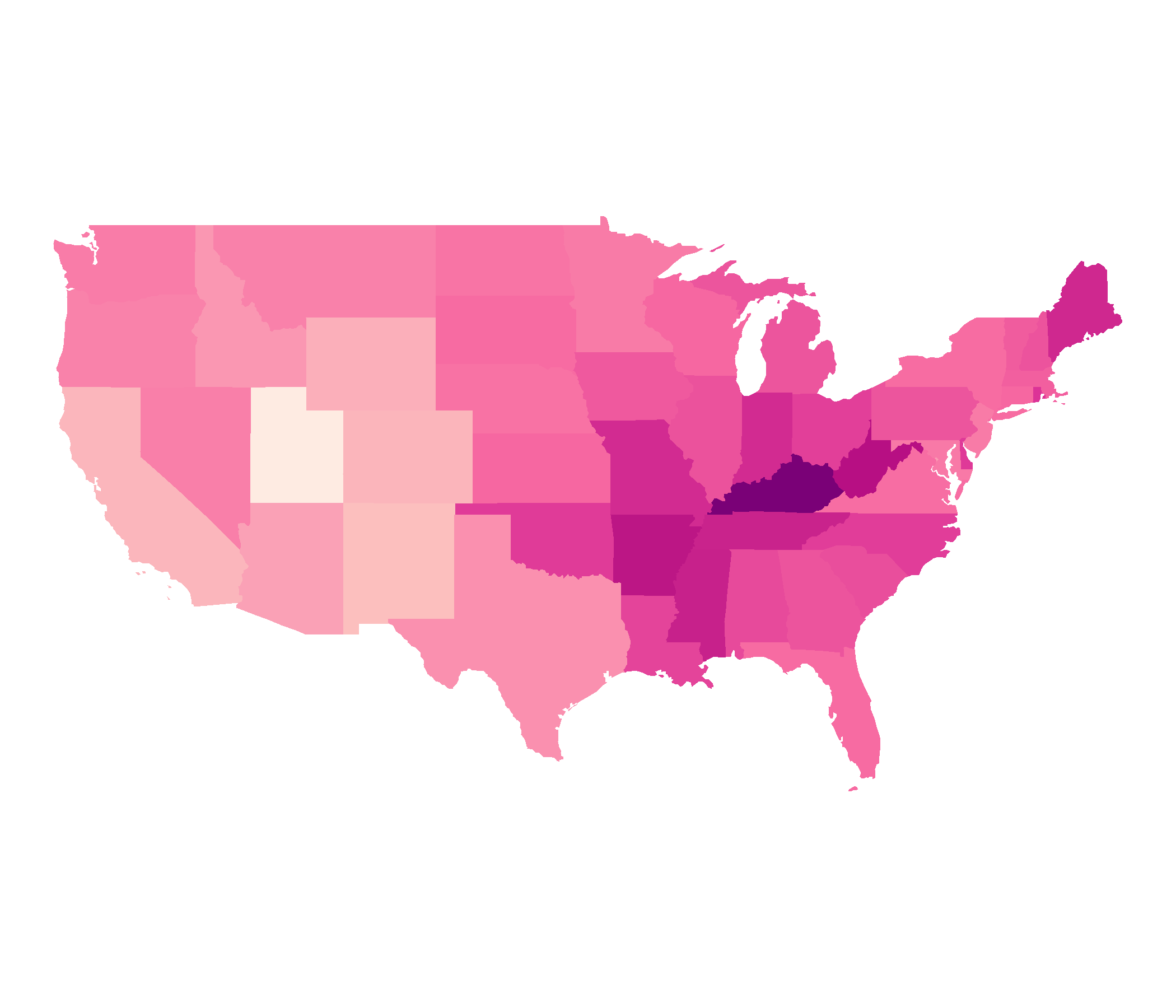
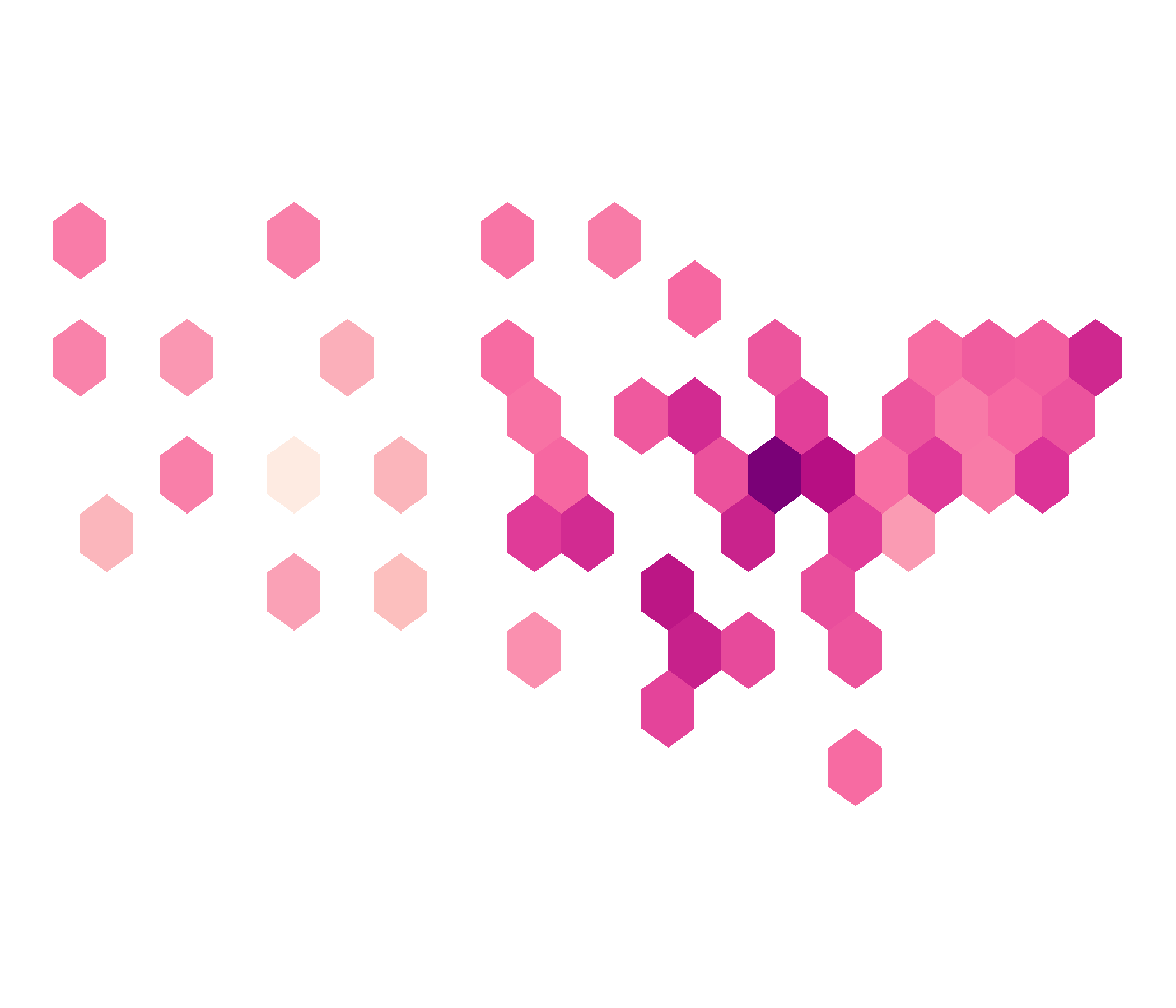
Non-Contiguous Cartogram
ncont <- cartogram_ncont(covid_districts_sl, weight = "population") %>% st_as_sf()ggplot(ncont) + geom_sf(data = covid_districts_sl) + geom_sf(aes(fill = Count), colour = NA) + scale_fill_distiller(type = "seq", palette = "RdPu", direction = 1) + theme_void() + guides(legend.position = "right")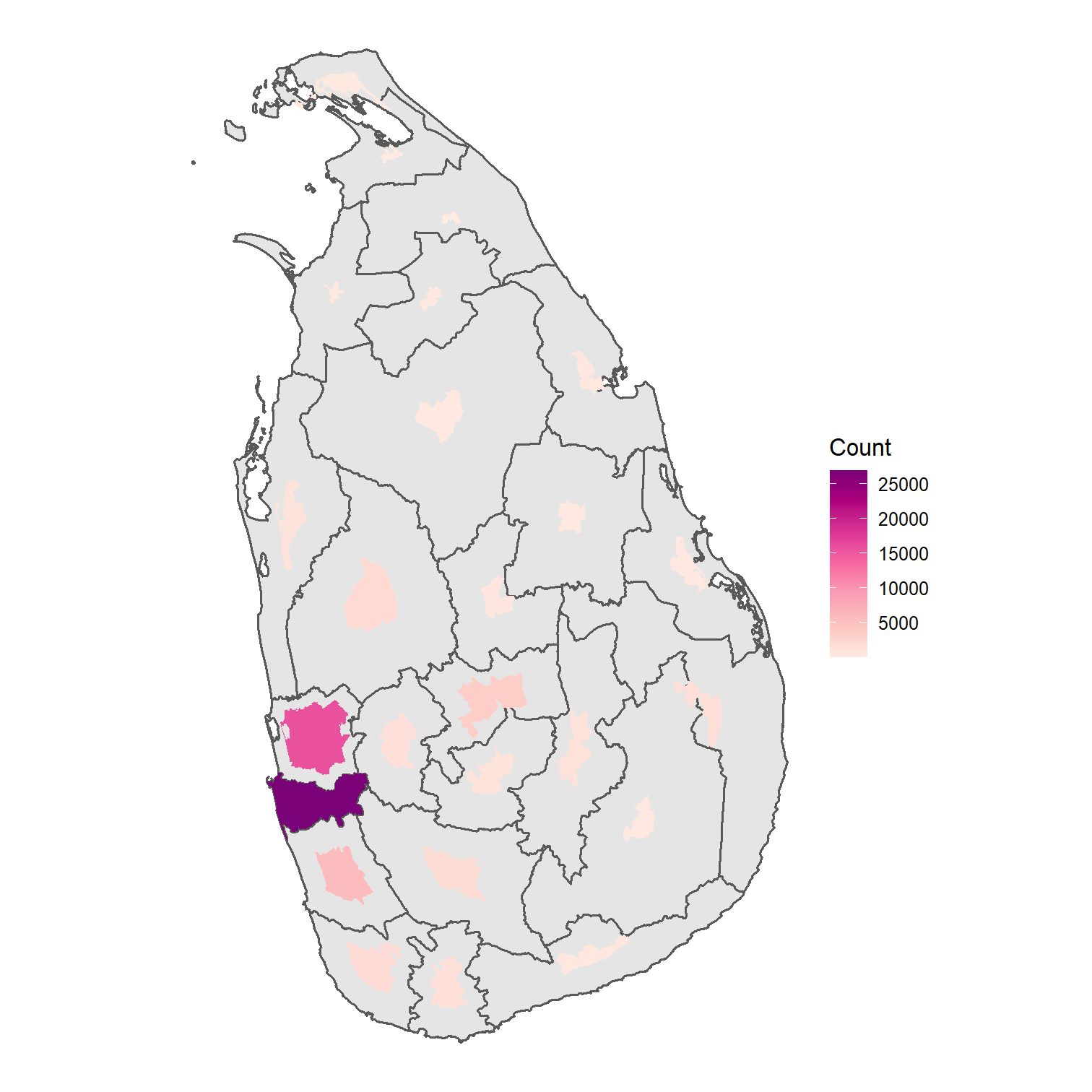
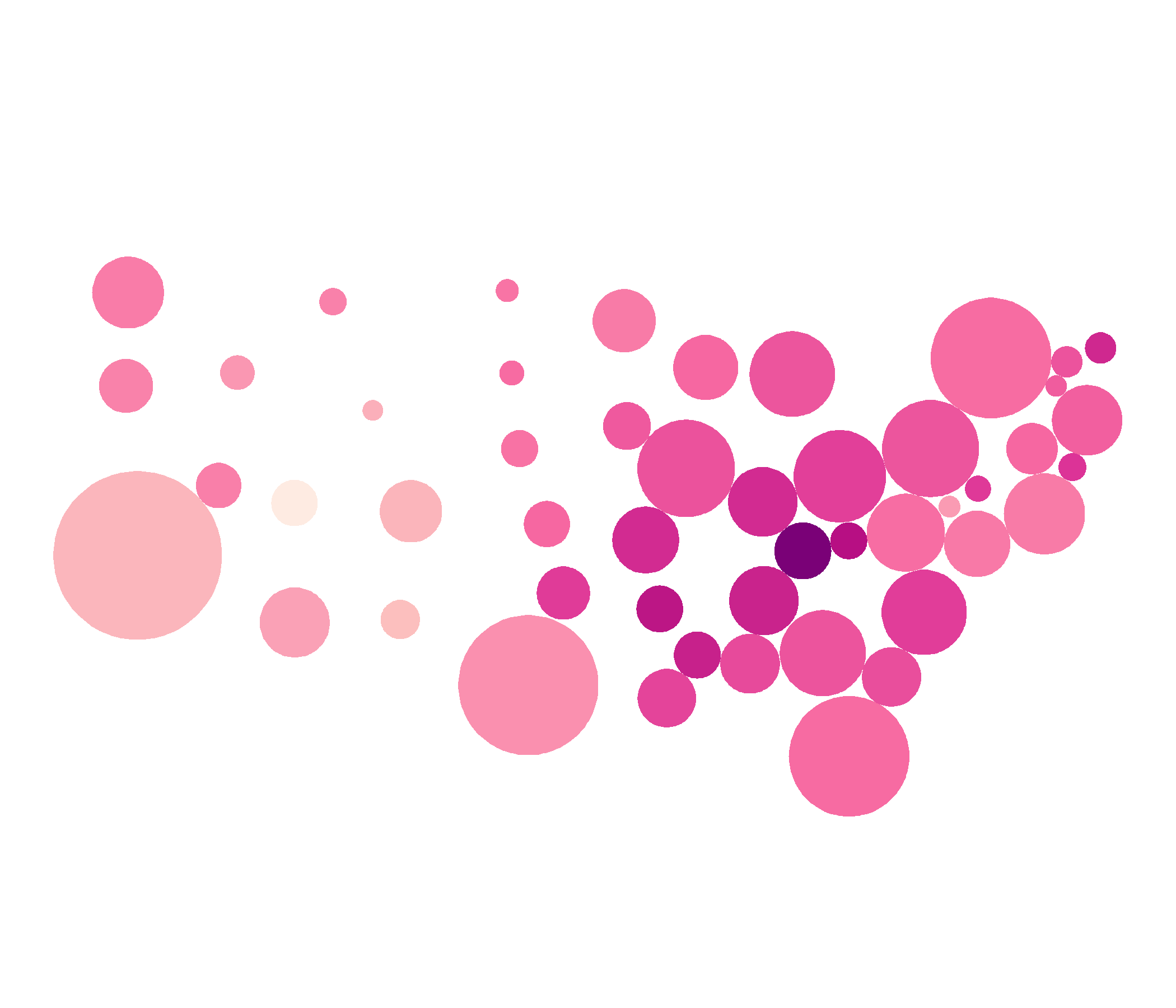
Dorling Cartogram
dorl <- cartogram_dorling(covid_districts_sl, weight = "population") %>% st_as_sf()ggplot(dorl) + geom_sf(data = covid_districts_sl) + geom_sf(aes(fill = Count), colour = NA) + scale_fill_distiller(type = "seq", palette = "RdPu", direction = 1) + theme_void() + guides(legend.position = "right")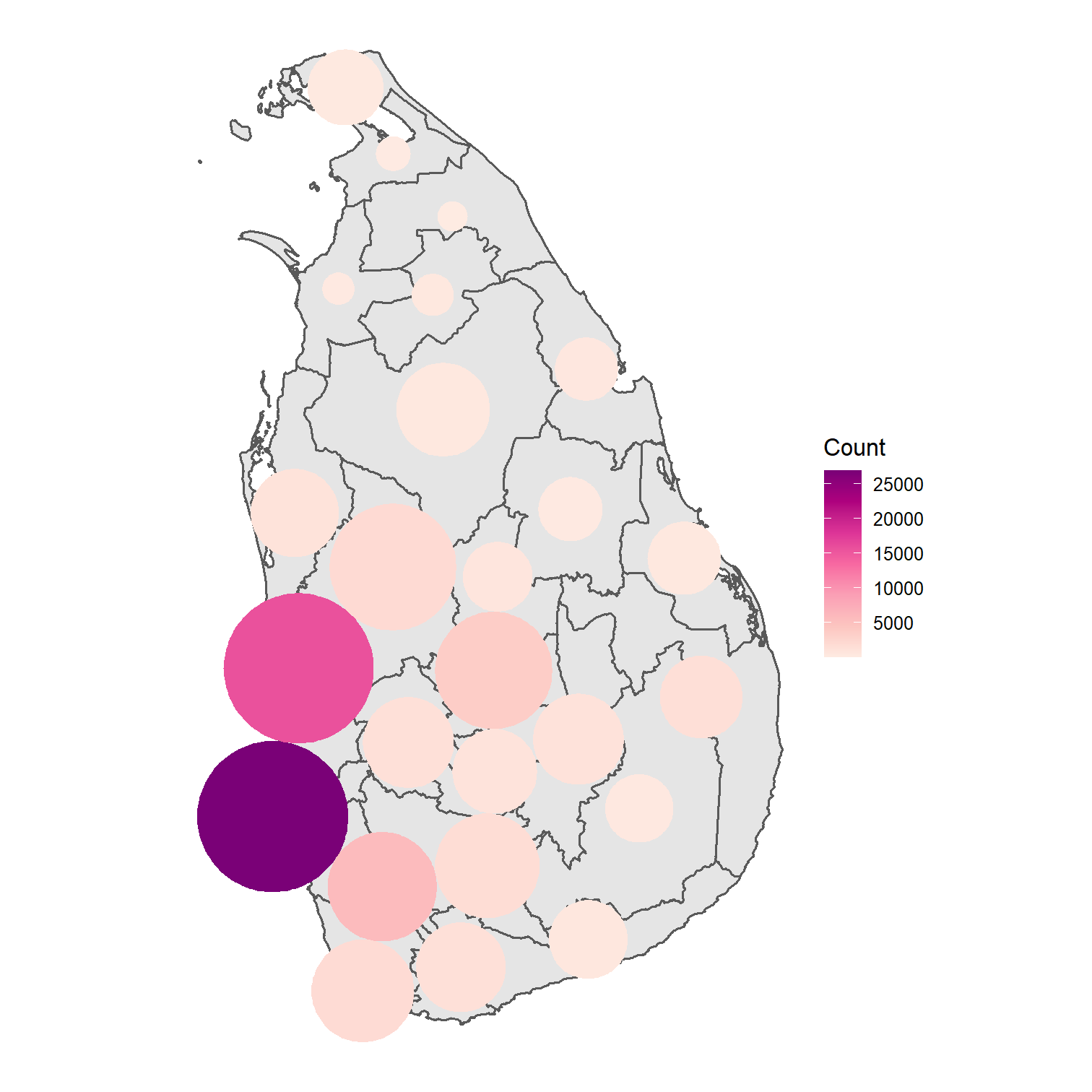

Ask me a question
Use the chat
Answer in discussion time
Thank you for listening!
Slides created via the R package xaringan.
The chakra comes from remark.js, knitr, and R Markdown.